Design and creation of a clinically compatible porcine donor for xenotransplantation
Wenning Qin1, Ranjith P. Anand1, Violette B. Paragas1, Jacob V. Layer1, Adam Akkad1, Matthew Pandelakis1, Shiyi Yin1, Sagar Chhangawala1, Robert A. Policastro1, Feng Li1, Xiaoyun Guo1, Yongqiang Xue1, Alexander K. Griffin1, Yinan Kan1, Russell J. Ernst1, Michele E. Youd2, David Heja2, Daniel J. Firl1, Susan C. Low1, Sierra Harken1, Tatsuo Kawai3, Takayuki Hirose3, Grace Lassiter3, Luis Queiroz1.
1Genome Engineering, eGenesis, Cambridge, MA, United States; 2Immunology, eGenesis, Cambridge, MA, United States; 3Center for Transplantation Sciences, Massachusetts General Hospital, Boston, MA, United States
Recent human decedent model studies and compassionate xenograft use explored the promise of porcine organs for human transplantation. However, the lack of a clinically compatible porcine donor is preventing xenotransplantation from advancing to clinical studies. Here we describe the design, creation, and long-term life-supporting function of transplanted kidneys from a genome-edited porcine donor in nonhuman primates.
The porcine donor was engineered to carry a total of 69 genomic edits that eliminate xenoantigens, overexpress human transgenes, and inactivate porcine endogenous retroviruses. Long-Read Whole Genome Sequencing revealed the presence of a single copy, intact transgenic construct at the AAVS1 genomic safe harbor site. In addition, Nanopore direct RNA sequencing showed transcription from all 3 cassettes (Figure 1), while single cell RNA sequencing demonstrated expression of human transgene mRNA in renal endothelial cells. At the protein level, immunohistochemistry detected human transgenic proteins in kidney cells. Expression was maintained among littermates of cloned donors, across cloning rounds, and during graft survival in the NHP model.
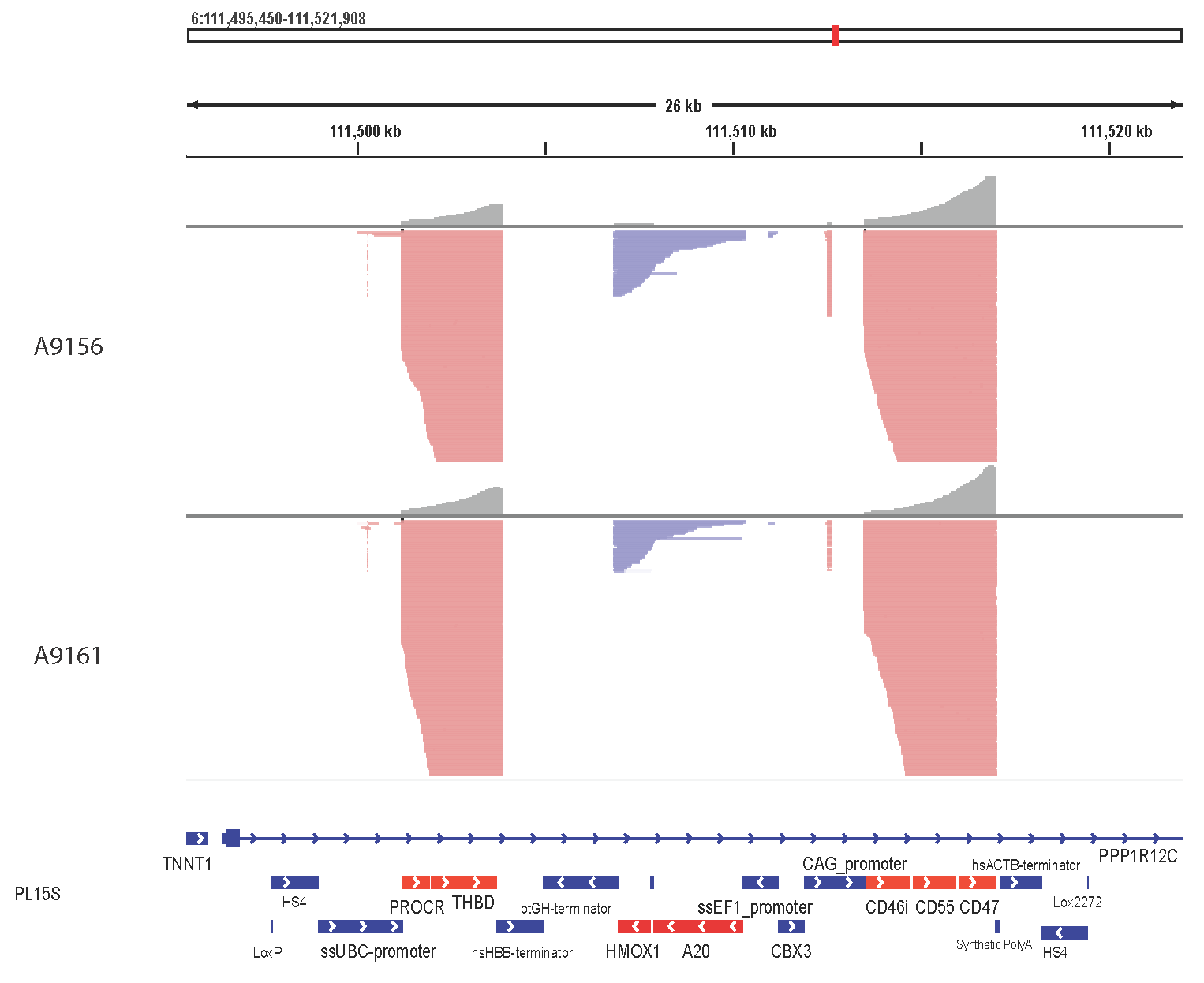
Figure 1: Nanopore direct RNA sequencing shows transcription from all 3 polycistronic cassettes, correct splicing from the 3 introns, and absence of aberrant splicing and transcription interference for all transcripts.
When transplanted into cynomolgus monkeys, long term graft survival and life supporting function were achieved (up to 758 days). Our study indicates that molecular incompatibilities between porcine donors and primate recipients can be overcome by selective gene edits. This success brings us closer to clinical studies of genome-edited porcine renal grafts.