Novel models for predicting the volume and the cell content of individual islets from 2D-microscopic projections for islet graft quality control
David Habart1, Barbora Radochova2, Martin Capek2,3, Sarah Suergiu1, Zuzana Berkova1, Eva Fabryova1, Klara Zacharovova1, Jiri Janacek2, Frantisek Saudek1.
1Laboratory for pancreatic islets, Institute for Clinical and Experimental Medicine (IKEM), Prague, Czech Republic; 2Laboratory of Biomathematics, Institute of Physiology CAS , Prague, Czech Republic; 3Light Microscopy, Institute of Molecular Genetics CAS, Prague, Czech Republic
Introduction: The correct islet graft dosing helps predict the transplantation outcome. The current quantification methods are based on the estimation of volumes of individual islets. The islet sizes and shapes are heterogeneous. The spherical islet model [1,2] presently used as the surrogate marker of the content of insulin secreting cells was challenged as grossly inaccurate [3]. Our long-term purpose is to establish an automated tool for islet graft quantification. To this end we are developing more accurate models for predicting the volumes as well as the cell content of individual islets from 2D-microscopic projections.
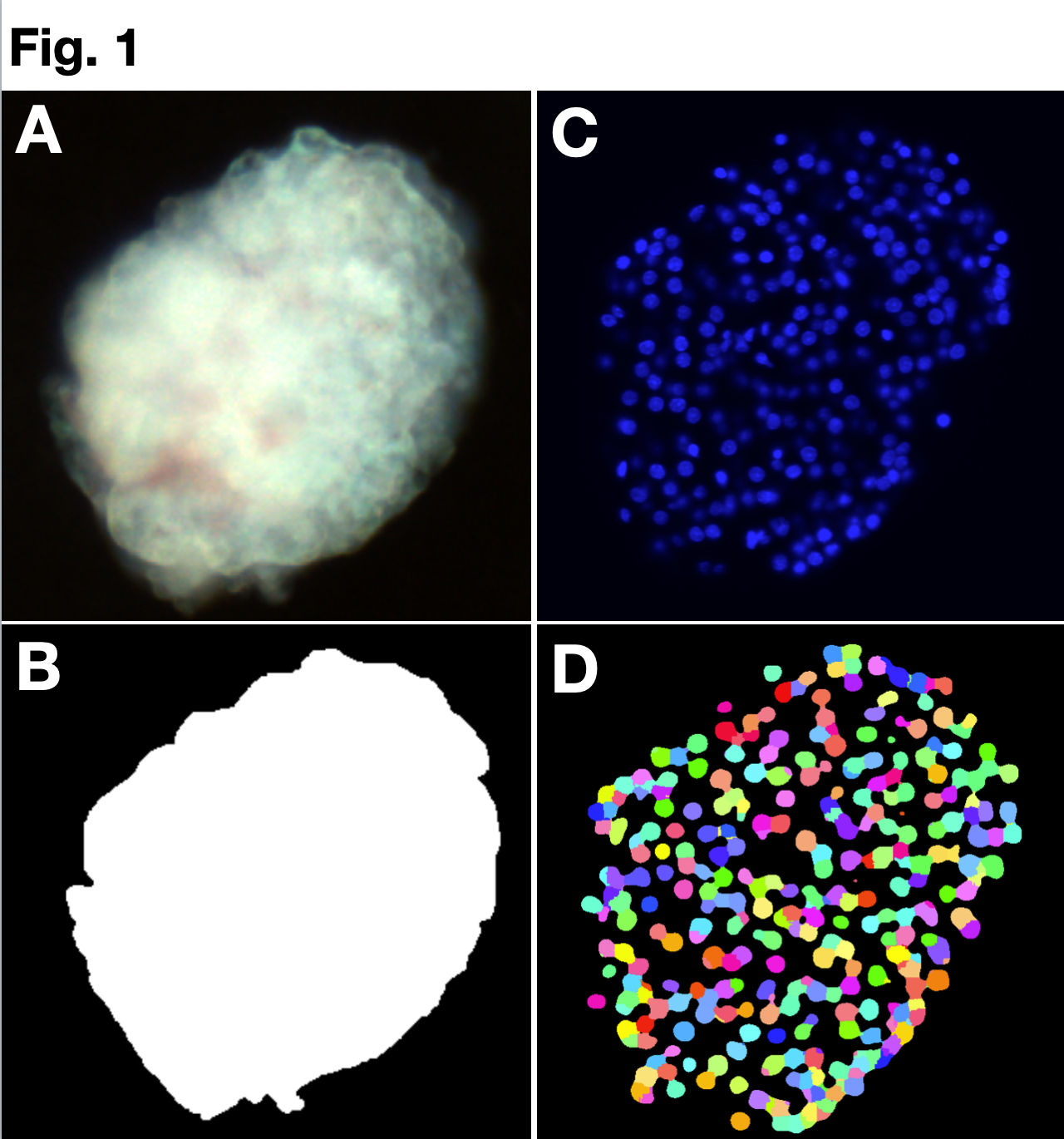
Methods: Our models are based on detailed observations of individual islets using the coupled 2D-and 3D-microscopy. High quality 2D-images of unstained islets (Fig. 1A) were acquired using Apotome microscope and manually segmented (Fig. 1B) and analyzed using the My-Segmentation tool in IsletNet.com. The Optical Projection Tomography (OPT) was used to collect 3D-images for the islet shapes and volume measurements. The Light Sheet Microscopy on cleared individual DAPI-stained islets (Fig. 1C) was used for the cell content quantification (Fig.1D). 3D-images were analyzed using Fakir (volume measurement) and Fiji macros (nuclear counting, Fig.1D) [4]. The islet cell content was validated by the determination of total DNA content in islet lysates using a fluorescence-based physical method (Qubit) and the genome size for cell number calculation.
Results: Our spinacle model for predicting the islet volume from 2D-projection was developed using 68 islets. The spinacle model was validated on 97 islets (size range: 83-546 µm, 8 donors), the volume of which was determined using OPT and Fakir software. The accuracy improvement using the spinacle model in comparison to the current standard method (spherical model) is shown in Fig. 2. Direct 3D-observation of the islet nuclei (49 islets, 4 donors, in blue) was in a good agreement with the islet DNA content (44 islets, 2 donors, in red), Fig.3. The results revealed that the current cell dissociation-based model [3] grossly underestimates the real islet cell content. The validation of our new proposed 2D-cell-count model on additional donors is underway.
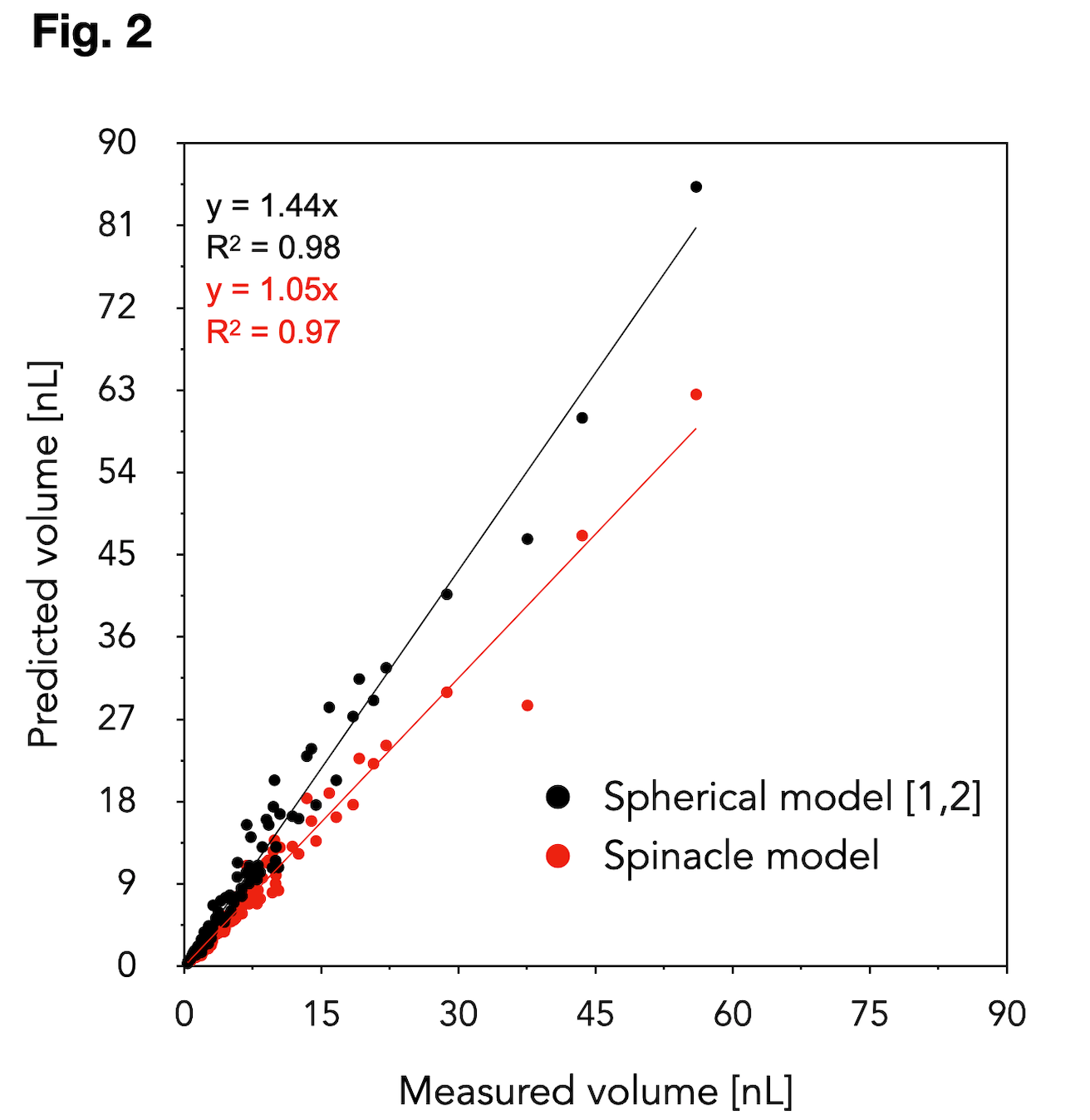
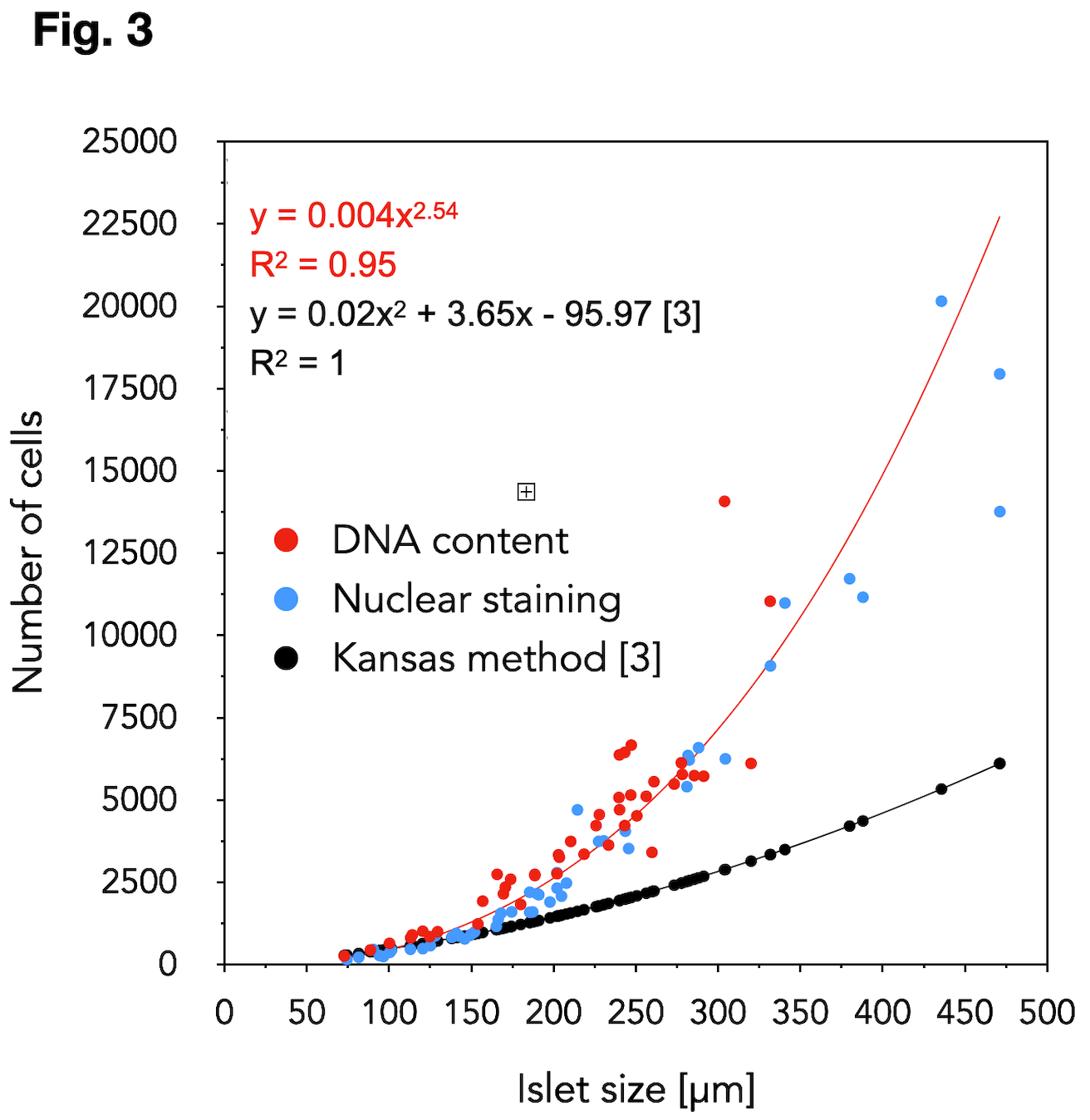
Conclusion: Our proposed models predict the islet volume and islet cell content more accurately than the current models. Their implementation into the web-service IsletNet is a step towards the cell-based graft quantification. Our findings are relevant for a fair comparison of the isolated vs. re-aggregated and stem cell-derived islets in the transplantation context.
This research was supported by grant NU22-01-00141 from Czech Health Research Council..
[1] Ricordi C, Pancreas. 1991; 2, 6: 242-244.
[2] Niclaus N, et al. Transplantation. 2008; 11, 86: 1603-1609.
[3] Ramachandran K, et al. Cell Transplantation. 2015; 7, 24: 1183-1194.
[4] Stringer C et al. Nature Methods. 2021; 1, 18: 100-106.