Immunotranscriptomic changes during two pig-to-human kidney xenotransplantation cases
Harvey Pass1, Chandra Goparaju1, Jeffrey Stern1, Karen Khalil1, Jacqueline Kim1, Bonnie Lonze1, Zoe A Stewart1, Adam Griesemer1, Elaina Weldon1, David Ayares2, Brendan Keating3, Robert A Montgomery1.
1NYU Langone Transplant Institute, New York, NY, United States; 2Revivicor, Inc., Blacksburg, VA, United States; 3Penn Transplant Institute, University of Pennsylvania, Philadelphia, PA, United States
Introduction: To determine whether subclinical early rejection-associated signals are reflected as biomarkers in blood cells during kidney xenotransplantation. We defined immunotranscriptomic changes before and up to 54 hours after kidney xenotransplantation in two brain-dead decedents.
Methods: Purified RNA was extracted from blood samples prior to xenotransplantation, and at 6, 12, 24, and 48 hours after xenograft reperfusion. Differentially expressed genes (DEG) between baseline and other timepoints were evaluated using two Nanostring panels that identify molecular phenotypes (mean of log2 expression for probes associated with target genes): the nCounter Human Organ Transplant Panel (HOT) and the nCounter Inflammation Panel. The HOT panel profiles 770 genes across 37 annotated pathways and the Inflammation Panel adds 255 additional targets. Panels were run on an nCounter Pro analysis system with analysis performed by nSolver. Fold change (FC) ratios compared to baseline gene expression were determined for all timepoints using Bonferroni tests. Immune cell phenotype as well as immune/inflammation pathways were determined.
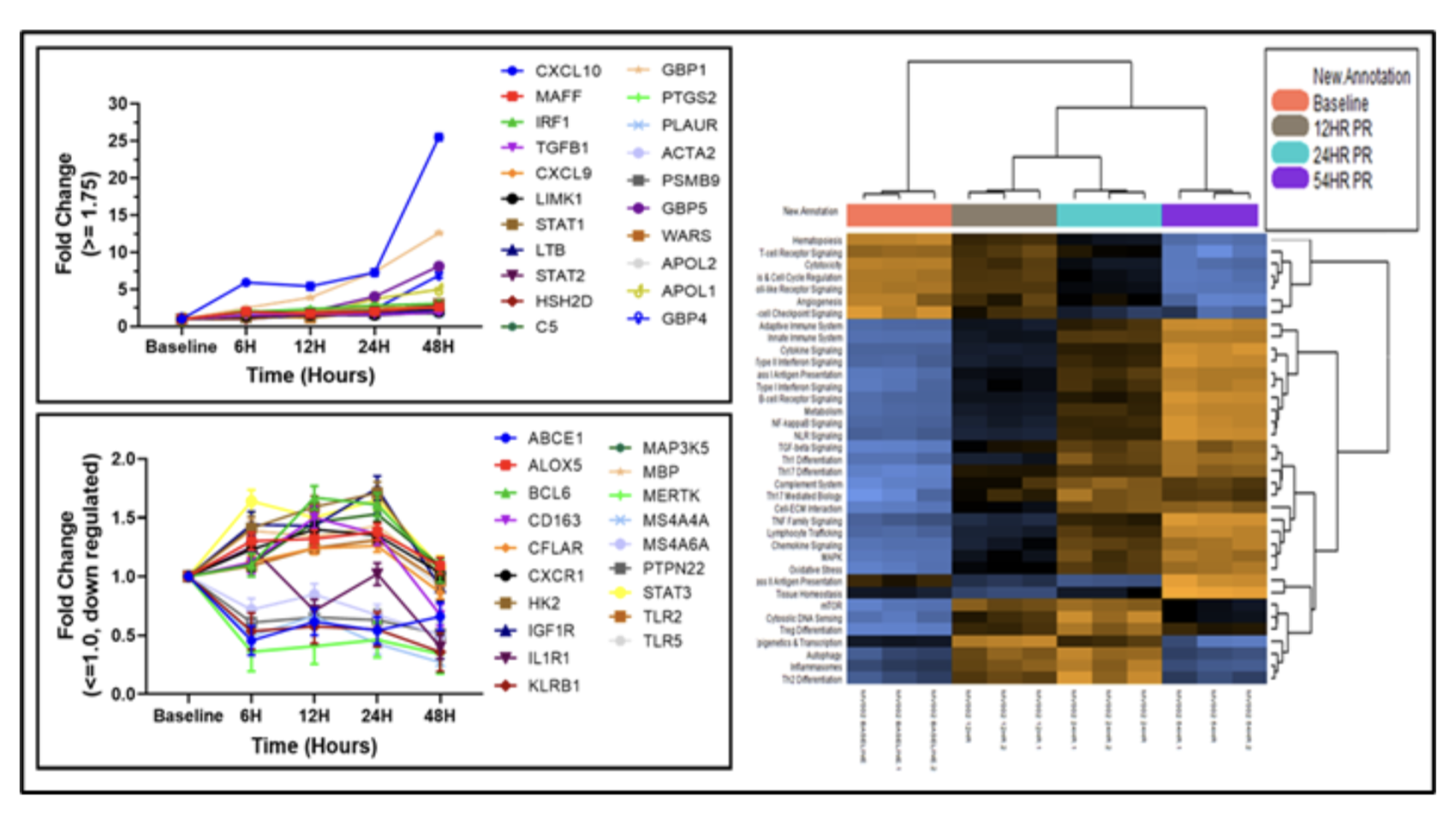
Results: Differentially expressed genes, which promote as well as attenuate rejection, were noted. Genes identified as biomarkers of rejection, graft outcome, and immunosuppression response included CXCL10 expression with a 25-fold higher at 48 hours compared to baseline (p<0.0001) along with GBP1 (FC=12.6, p<0.0001) and GBP4 (6.89 FC, p<0.0001). Conversely, CD163, a macrophage marker associated with chronic allograft function, decreased at 48 hours (FC= 0.673, p<0.0001), as did MS4A6A (FC= 0.503, p<0.0001), which is elevated after complement deposition and accumulation of natural killer cells and monocytes/macrophages in capillaries. Macrophage, T-cell, cytotoxic T cell, and CD45 scores decreased significantly at 48 hours. Pathway analyses were notable for decreased T-cell receptor, check point signaling, Toll-like receptor signaling, and angiogenesis at 48 hours along with increased cytokine, interferon and chemokine signaling.
Conclusion: This characterization of early transcriptomic changes after kidney xenotransplantation illustrates an early balancing of rejection mechanisms by suppressive pathways which are stimulated either intrinsically or through exogenous immunosuppressive drug treatment.
Supported by Lung Biotechnology, a wholly owned subsidiary of United Therapeutics..