Defining the human anti-pig xenoreactive T cell repertoire
Farshid Fathi1, Nathan Suek1, Christopher Parks1, Megan Sykes1,2,3.
1Columbia Center for Translational Immunology, Department of Medicine, Columbia University Medical Center, New York city, NY, United States; 2Department of Surgery, Columbia University Medical Center, New York city, NY, United States; 3Department of Microbiology and Immunology, Columbia University Medical Center, New York city, NY, United States
Introduction: Xenotransplantation from pigs is a promising solution to address the organ shortage and preventing T cell-mediated rejection will be important to realizing its potential. The human anti-pig xenoreactive T cell repertoire and characteristics have not been analyzed in detail. In this study, we sought to characterize human anti-pig xenoreactive T cell responses using single cell RNA sequencing combined with TCR sequencing(1).
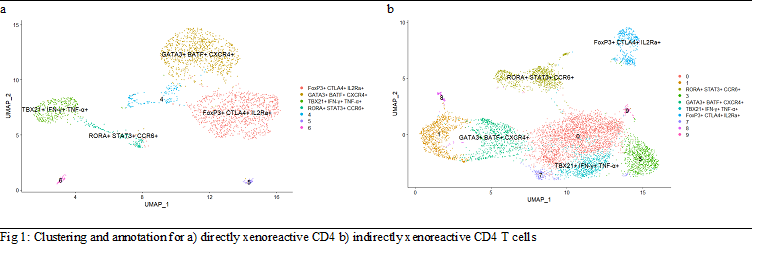
|
TBX21hi IFN-γhi TNF-αhi (Th1) |
GATA3hi BATFhi CXCR4hi (Th2) |
RORAhi STAT3hi CCR6hi (Th17) |
FOXP3hi CTLA4hi IL2Rahi (Treg) |
|
|
Direct |
12%, |
34% |
5% |
38% |
|
Indirect |
9% |
10% |
11% |
5% |
Methods: We established human-anti-pig mixed lymphocyte reactions (MLRs) with pig antigen presentation by the direct and indirect pathways. For the direct pathway, pig DCs were differentiated from PBMCs of SLAHH pigs. As responders, human T cells were MACS purified and labelled with Violet Dye. For the indirect pathway, pig PBMCs were irradiated at 100 Gy to generate apoptotic cells and combined with human macrophages differentiated from a healthy donor PBMCs. After overnight incubation, cells were CD14+ MACS-enriched before co-culture with purified T cells from the same human donor for 6 days. From each pathway, 17000 Violet Dye low human anti-pig xenoreactive CD4 or CD8 T cells from each response were separately FACS-sorted, combined with 10x Chromium 5’ RT reagents, and loaded onto a Chromium microfluidics chip and controller for single cell/10X barcoded primer bead droplet formation. Full-length cDNAs were synthesized from polyadenylated mRNAs and labeled with a unique 10x barcode. For 5’ gene expression sequencing (5’GEX-seq), cDNAs were amplified followed by sequencing libraries preparation and sequencing on an Ilumina NovaSeq 6000 Sequencer (CUIMC Genomics Core). For TCR-seq, target enrichment was performed using TCR-specific outer and inner primers, followed by library preparation and sequencing on an Ilumina NextSeq 550 Sequencer (CUIMC Genomics Core). FASTQ files were processed using the 10x Genomics Cell Ranger 3.1.0 with the GRCh38-3.0.0 reference. Resulting scRNA-seq data was further processed and analyzed using the Seurat v3 pipeline.
Results: From the direct pathway, we sequenced 2477 CD4 cells and 103 CD8 cells. From the indirect pathway, we sequenced 7535 CD4 cells. Transcriptionally-defined functional T cell types are presented in Figure 1 and Table 1. TCR diversity was assessed by Shannon’s clonality (from 0 to 1, with higher scores indicating less diversity). For indirect CD4 TCRβ, clonality was 0.073. For direct CD4 TCRβ, clonality was 0.029. Immunodominant clones were detected in both responses, with top frequencies of 2% and 1.9% for direct and indirect responses, respectively.
Conclusion: By combining single cell RNA and TCR sequencing, we have demonstrated that direct and indirect CD4 xenoresponses include all classes of CD4 helper responses and possibly Tregs. Both responses involve diverse TCR repertoires, though some immunodominant clones are identified in each.
[1] Sykes M, Sachs DH. Progress in xenotransplantation: overcoming immune barriers. Nature Reviews Nephrology. 2022 Dec;18(12):745-61.