The tissue common response module (tCRM) score predicts treatment response for acute cellular rejection following pancreas transplant
Audrey E Brown3, Yvonne M Kelly3, Arya Zarinsefat1, Worner Giulia1, Raphael PH Meier2, Minnie M Sarwal1, Andrew Posselt1, Zoltan G Laszik4, Tara Sigdel1, Peter Stock1.
1Transplant Surgery, University of California, San Francisco, San Francisco, CA, United States; 2Transplant Surgery, University of Maryland, Baltimore, MD, United States; 3General Surgery, University of California, San Francisco, San Franscisco, CA, United States; 4Pathology, University of California, San Francisco, San Francisco, CA, United States
Introduction: The tissue Common Response Module (tCRM) score, which is based on the expression levels of 11 genes (BASP1, ISG20, PSMB9,RUNX3, TAP1, NKG7, LCK, INPP5D, CXCL9, CD6, CXCL10), was developed using genomics data from heart, liver, lung, and kidney tissue with acute cellular rejection (ACR). Using our institution’s repository of pancreas transplant specimens, we sought to validate the tCRM score in pancreas samples with ACR and to determine whether it correlates with treatment response.
Methods: Fifty one formalin fixed paraffin embedded pancreas biopsy specimens: 14 normal, 14 Grade 1 (G1) ACR, 14 Grade 2 (G2) ACR, and 9 Grade 3 (G3) ACR were analysed using multiplex RNA sequencing. Differential gene expression and pathway analyses were performed using a panel of 804 unique genes, including the genes that make up the tCRM score. Whole transcriptome spatial RNA analysis is ongoing to further investigate patterns of gene expression in distinct regions and cell populations.
Results: Significant differences in gene expression were seen in G2 and G3 ACR, but not G1 ACR, when compared with normal pancreas samples. Pathway analyses demonstrated that the genes enriched in G2 and G3 ACR were associated with inflammatory pathways like interferon signalling, antigen processing and presentation, and phagocytosis. A statistically significant difference was found in the tCRM scores for G2 ACR and G3 ACR, when compared to the tCRM score for normal pancreas tissue. Patients with rejection that was resistant to treatment (defined as recurrent rejection within 2 months of the initial rejection episode) had significantly higher tCRM scores (Figure 1). This difference remained significant for patients with G1 ACR, but was not significant for patients with G2 or G3 ACR (Figure 2).

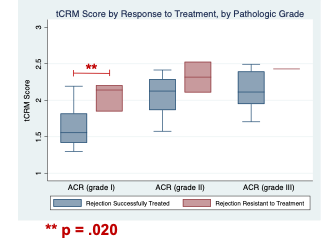
Conclusions: We found that the tCRM score not only correlates with commonly used histologic grading criteria, but also identifies patients with ACR who are at risk for treatment resistant rejection and thus may warrant more aggressive initial treatment with lymphodepleting agents. Further work using spatial transcriptomics, including whole transcriptome differential expression analysis and spatial deconvolution is underway to better elucidate the regions and cell populations involved in the development of acute pancreas rejection.
Audrey Brown was supported by the UCSF National Institutes of Health (NIH) FAVOR T32 Training Grant, grant number 2T32AI125222-06A1..