Integrative omic profiling and analyses in two pig heart to human xenotransplants
Brendan Keating1, Eloi Schmauch2, Brian Piening3, Bo Xia4, Chenchen Zhu5, Boa-Li Chang1, Karen Khalil4, Jacqueline Kim4, Elaina Weldon4, Harvey Pass4, David Ayares6, Adam Griesemer4, Massimo Mangiola4, Jeffrey Stern4, Michael P Snyder5, Jef Boeke4, Robert A Montgomery4.
1Penn Transplant Institute, University of Pennsylvania, Philadelphia, PA, United States; 2Massachusetts Institute of Technology, Cambridge, MA, United States; 3Earle A. Chiles Research Institute, Providence Cancer Center, Portland, OR, United States; 4NYU Langone Transplant Institute, New York, NY, United States; 5Department of Genetics, Stanford University, Palo Alto, CA, United States; 6Revivicor, Inc., Blacksburg, VA, United States
Introduction: Pig organs offer significant advantages for transplant into humans, and may alleviate the current critical shortage of suitable organs. Advances in genetic knockout models of key xeno-antigens such as α-1,3-Gal and ethical research using recently deceased brain-dead human donors has enabled the first sets of xenotransplants (XTx) to be performed. In the Summer of 2022 two pig heart xenografts were transplanted into two human decedents in NYU for ~3 days, with the primary aims being to assess hyper-acute rejection and appropriate xenograft functioning. We performed multi-omic profiling to assess potential xenogenic interactions between pig and human genomes, and dynamic biological interactions across blood samples to assess immunological and other physiological changes.
Methods: Short-read whole-genome sequencing (WGS) was performed across the human and pig genomes. Bulk FFPE RNAseq in peripheral blood mononuclear cells (PBMCs) and biopsy tissues, and Single-cell RNA-sequencing (scRNAseq) of PBMCs were performed every 6 hours on up to 25 timepoints across the two xenoheart procedures. Proteomics in plasma was performed using liquid chromatography mass spec (LC-MS), PECAN, SWATH-MS. Metabolomics was performed using gas chromatography (GC)-MS & LC-MS. Conventional, digital and electron microscopy imaging of 30 biopsies was also performed.
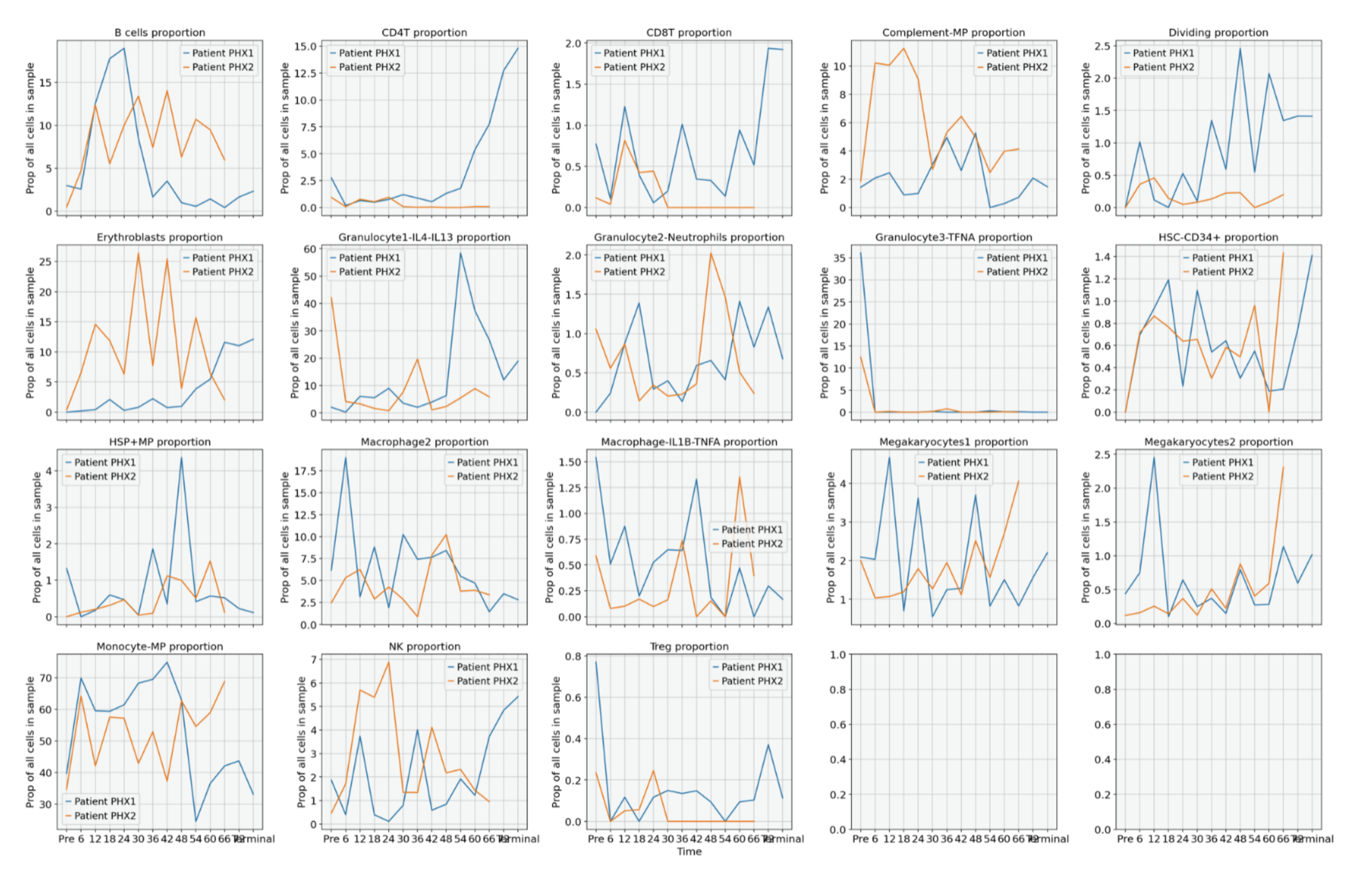
Results: Multi-omic profiling of blood from two pig heart to human xenotransplants shows significant changes increases in metabolic and antigen presentation pathways after the 40 hours timepoints in the first xenoheart decedent (PHX1 in Figure 1). This timeframe coincides with significant physiological changes including major increases in liver transaminase enzyme activity. Significant proteomic and metabolomic pathway changes were observed including Glycolysis/Gluconeogenesis metabolism (p< 5 x 10-20), Pyruvate metabolism (p < 7 x 10-11) and antigen processing and presentation (p = 5.6 x 10-5). Utilizing PBMC scRNAseq across the same time points, we identify key changes in immune cell types and subtypes, including T-cell subtypes, B cells, granulocytes, macrophages, and monocytes. Notably we see a particularly sharp increase in CD4 T-cells, followed by CD8 T-cells with these changes preceded by a marked increase in B cells, whose timepoint-associated phenotypes exhibit IL1B- & IL17-related activation at 6 and 12 hours, followed by B-cell proliferation and BCR activation at 18 and 24 hours.
Conclusion: Multi-omic profiling including scRNAseq profiling in two pig to human xeno heart-transplant decedent datasets show significant changes in metabolic and immune function after 40 hours in the first decedent subject.