Duke University
A histocompatibility epitope prediction software for xenotransplantation donor-recipient optimization
Joe Ladowski1, Henry Chapman1, Imran J Anwar1, Isabel DeLaura1, Annette Jackson1, Stuart J Knechtle1, Bruce Rogers1, Jean Kwun1.
1Duke University, Department of Transplant Surgery, Durham, NC, United States
Background: Antibody-mediated rejection remains the barrier to long-term success in preclinical models of xenotransplantation. It is well-known in human renal allotransplantation that donor-recipient MHC mismatches are associated with the development of donor-specific antibody (DSA) and worse graft outcomes. While allotransplantation has a variety of tools available (e.g. HLA Matchmaker, HLA-EMMA, etc). There are currently no available similar software programs for studying donor-recipient MHC compatibility in xenotransplantation.
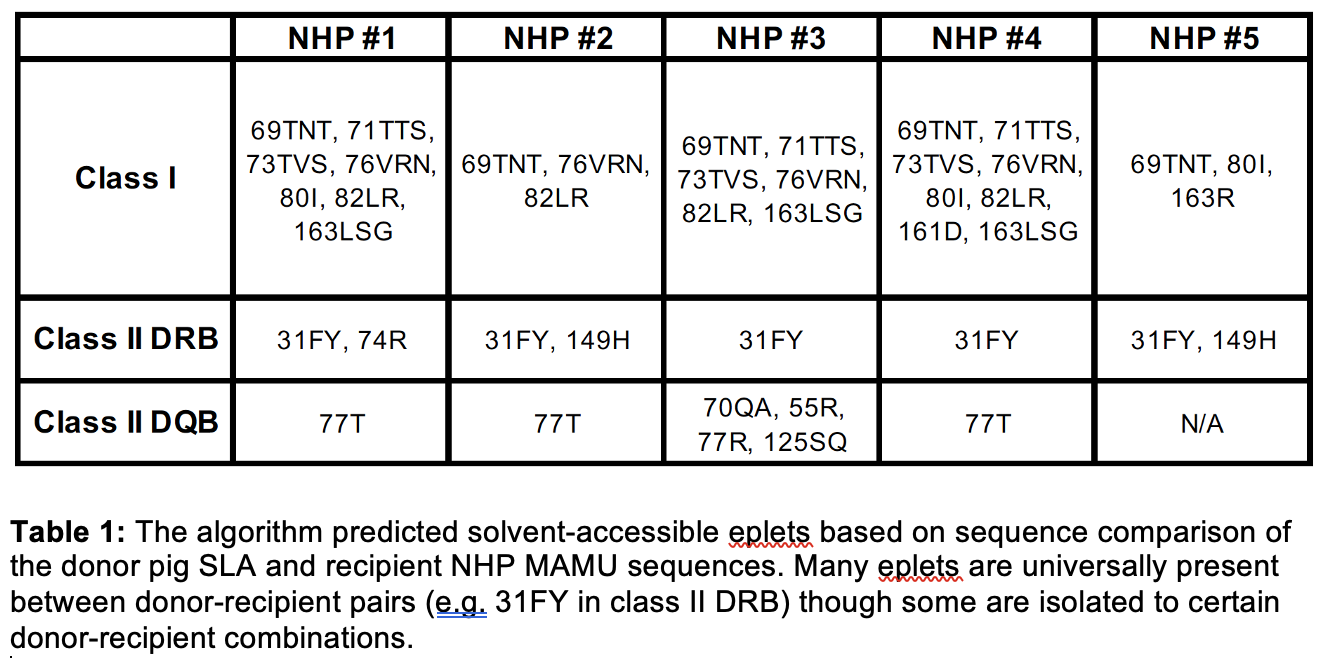
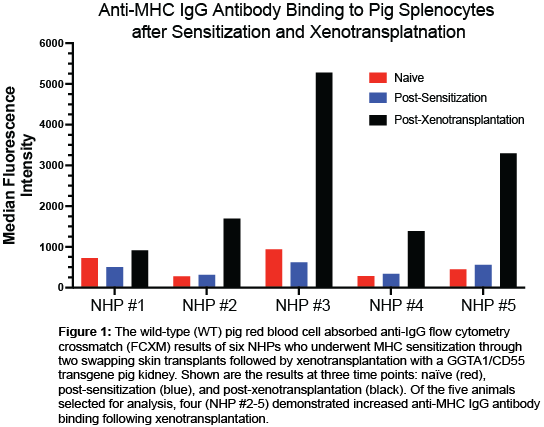
Methods: Six non-human primates (NHP) rhesus macaques were chosen for analysis. Briefly, the NHPs underwent two swapping skin transplants to induce MHC sensitization then bilateral nephrectomy and kidney transplantation from an aGal knockout, human CD55 transgenic (GGTA1/CD55) pig (National Swine Resource and Research Center, Columbia, MO). To study the anti-MHC donor-specific antibodies in the sera, cryopreserved sera was glycan-absorbed on wild-type (WT) pig red blood cells (RBCs) at three time points: prior to sensitization, post-sensitization, and at the time of euthanization post-xenotransplantation. A flow cytometry crossmatch with the glycan-absorbed sera was performed on donor pig splenocytes to evaluate anti-IgG antibody binding to pig cells. The donor pig SLA and recipient NHP MAMU sequences were using the MHC Matchmaker algorithm, with specific attention at exons 2 and 3 of classical class I, -DRB, and -DQB loci. NetSurfP-3.0, a structural prediction program, was used to predict the solvent accessibility and the associated amino acid sites were correlated to known antibody-verified HLA eplets.
Results: One animal was excluded from analysis based on technical complications post-xenotransplantation. The FCXM demonstrated 4 of the 5 (80%) of the NHPs demonstrated increased anti-MHC IgG antibody binding post-xenotransplantation (Figure 1). The MHC Matchmaker program prediction of solvent-accessible eplets between the donor pig and recipient NHP are shown in Table 1.
Conclusions: We describe the first version of a histocompatibility software program to compare the MHC amino acid differences between and across non-human species. Validation experiments consisting of antibody binding-elution and MHC mutagenesis are ongoing, but this algorithm could potentially be applied to a) non-human primate allotransplantation b) preclinical xenotransplantation c) clinical xenotransplantation.
Portions of the xenotransplantation work performed at Duke University is in collaboration with eGenesis . JML is funded by the Duke Resident Physician-Scientist Program – NIAID (1R38AI140297) .
Lectures by Joe Ladowski
| When | Session | Talk Title | Room |
|---|---|---|---|
|
Sat-28 10:00 - 11:30 |
Immune monitoring for rejection / Immunomodulation / tolerance | A Histocompatibility Epitope Prediction Software for Xenotransplantation Donor-Recipient Optimization | Indigo 204 |
